| Organism | |||
| GO term(s) | Biological Process: GO: 0006810 |
||
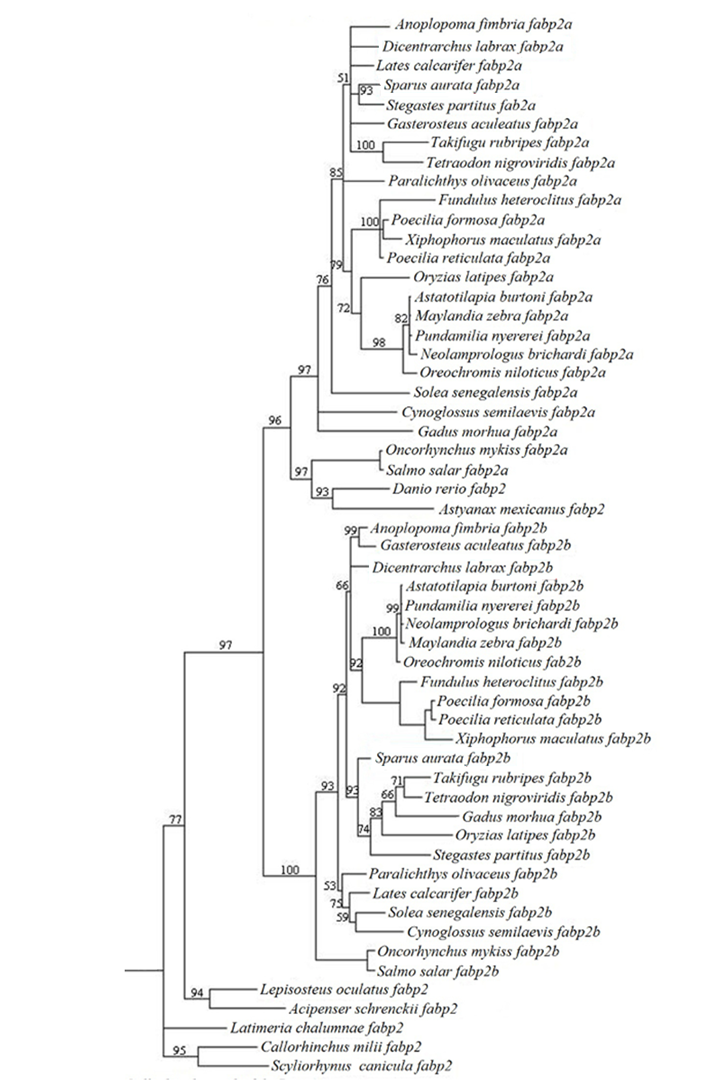
| Phylogeny | |||
| Genomic Map Locations | JH556843.1 fabp2b JH557230.1 |
||
| Paralogs | |||
| Sequencing Methods | fabp2b: NCBI: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NW_004088019.1) annotated using gene prediction method: Gnomon, supported by EST evidence. |
||
| Citations | |||
| Sequence | >XM_005807140.1 Xiphophorus maculatus fabp2a TTTAAACGGCCACATCATCACGCAGATGACGAGATAAGAGCGTACGCAGCTTAAAAGGAGCGCCGCACTCGCAGTAGGAC GGTCCTTTCTGCACAGTGGTTCAGCTCGCCTCAAGCAGTCAACATGACCTTCAACGGCACCTGGAAGGTGGATCGCAATG AAAACTATGAGAAGTTTATGGAACAAATGGGAGTAAACATGGTGAAGAGGAAGCTGGCCTCTCATGACAACCTGAAGATC ACCATCGAACAGACTGGAGACAAGTTTCATGTGAAGGAGAGCAGCAATTTCCGCAACATTGAAATAAACTTCACCCTGGG AGTCAACTTTGAGTACAGCCTGGCAGATGGAACACAACTATCAGGTGCCTGGACCATGGAGGGTGACACTCTTAAGGGGG TTTTCAACAGGAAGGACAACGGAAAACCGCTGACAACAGTCAGAACCATCCAAGGAGACGAACTCGTTCAGACCTACAAC TACGAAGGTGTGGATGCAAAGAGGATTTTCAAGAGAAGTTAGACCCGTCTTCTTTACTAAACTAAGTGAATTATGAAATC ATGTACAGTATTGTGTTGAACTGTCACATACATTCAAAAGTGCCAGATTTGTGATGCATTTTTATACCTGCATTAATAAA ATTAGAACAGCCGG
>XM_005813397.1 Xiphophorus maculatus fabp2b GGTGAGGGTGAGAGTGGAGGGTGGGAGTTGACAGCGACATGGGGATGGGGTGGGTATAAATGAGGGCCAGTGGGCATTCT CAGTCACACGGTTGTCCCCAGTGCCTTGTAACCAGCAGAGACGCTTGCAGCCATGGCTTTCAATGGGACCTGGAAGATTG ACCGCAATGAAAACTACGACAAGTTCATGGAGCAGCTGGGTGTCAACGTGGTGAAACGCAAGCTGGGTGAGCACGACAAC CTGAAGCTCACTTTTGAGCAAAATGGGGACAAGTTTCACATCAAAGAGTCCAGCACGTTCCGCACCAAAGAGATCGACTT CACCCTGGGCGTTCAGTTTGACTACTCTATGGCTGATGGAACCGACCTCGTAGGCACATGGGTGCTAGAGGGTGATACGC TGAAAGGGACTTTCAACAGGAAAGATAACAACAAGCTCCTGATCACCACAAGATCACTGGTTGGAGGAGAGCTTGTGCAG AGTTACAGCTATGAAGGCGTGGACGCAAAACGAATTTTCAAGAAGCAGTAACCAGGATCCAGCTGTTTATACTTGTACTT TTATATAGCTCACACAACTTTCTTTTCATCTTAACATTAGAGAACAACCAACCCTAAGGTGTATTCTCAACTCATTCATT TTATCTACCTGAAAATGTTTTTATATATATGTACAAATGCTTTTGGGGATGAATCATTGTAAAGGTGCTGTACTGCCAAT ACTTGAGTGAAACAATAAACAGAAAGGTGTTGCCCCCTGACTTGACAACTGATGACCTTGACAGACACATTTTGTACTCC GTGTCCAGTTCTCAGGCCAGGTCAGGAAGTCTAAATAAAGGGAGCTAAAAGAGCTGTTCCAACTGCAAA
|
||||||||||