| Organism | |||
| GO term(s) | Biological Process: GO: 0006810 |
||
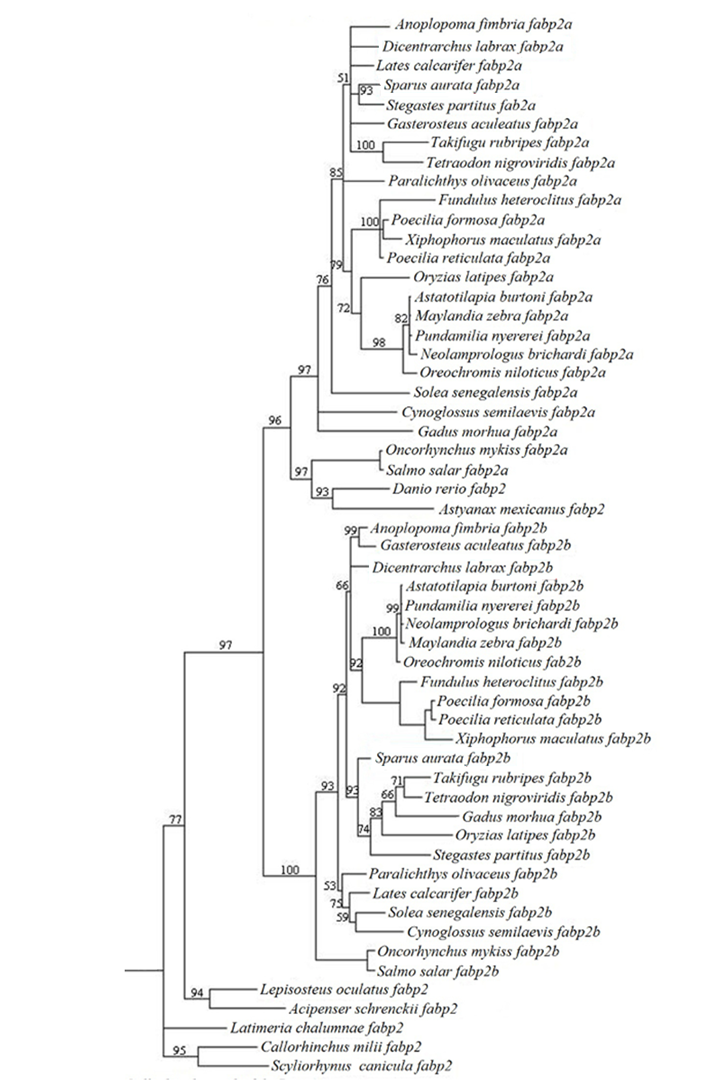
| Phylogeny | |||
| Genomic Map Locations | LG6 fabp2b LG3 |
||
| Paralogs | |||
| Sequencing Methods | fabp2b: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NT_167465.2) annotated using gene prediction method: Gnomon, supported by EST evidence. |
||
| Citations | |||
| Sequence | >XM_003452249.2 Oreochromis niloticus fabp2a GAGATAGGAGGGTGTGTGGTTTAAAAGGAGCGGCAGAGTTTGAAAAGGGCACTCCTTGCTGCAGAGTCCTCCAGCTCACC TCACCGCCACCATGACTTTCAACGGCACTTGGAAAGTTACTCGAAATGAGAACTATGACAAATTCATGGAAAAAATGGGA GTTAACATGGTGAAGAGGAAGCTGGCTGCTCACGACAACCTCAAGATTACCATCACGCAGGAGGGAGACAAGTTTAACGT CAAGGAGAGCAGCACTTTCCGAAGCATCGAAATTGACTTTACTCTCGGAGTTACCTTTGAGTACGCCCTTGCAGACGGAA CAGAACTATCAGGTGCGTGGACCTTGGAGGGAGACACGTTGAAGGGAATTTTCAAGAGAAAGGACAACGGAAAAGAGCTG ACGACAACCAGAATAGTTCAAGGAGATGAACTCATACAGAGTTACAGCTACGAAGGTGTGGATGCAAAGAGGATTTTCAA GAGGAGTTAGACCAAATCCATCTAAAGAACCGAAGTGAAATATTTTCAGGGTTGCATACAGTTTTGTGTCAAGTCATCAA CAACATGTAATTCATCATATTTTGATAGCTGCATTAATAAAGTCACAACTGCCTATTAA
>XM_003450322.3 Oreochromis niloticus fabp2b GCGTATCCTTGAGGTCGTGCAGCTATCGGCCTACATTTGACACCGTCTTTAAATGGCCACATCAGCAGACAGATAATCAG TGAGATAAAGAACAGCATCTTCCATGCCAGCTCAAGGAGGGGTGGATGTGTATAAATTGAGGCCTTTGGGGAATCACAAT CTCACGGTCTGCTGTAGCGCTTCATCATCAGCTGCCTTCAACCCGACGAGCCTGCAGCCATGGCCTTTAATGGAACCTGG AAGGTCGACCGCAGTGAGAACTATGTCAAGTTCATGGAGCAAATGGGTGTTAATCTTGTAAAACGTAAGGTGGCAGAGCA CGACAACCTGAAGATCACCATCGAGCAGAACGGGGACAAGTTTCACGTCAAAGAGTCCAGCACTTTACGCACCAAAGAGA TTGACTTCACTCTGGGCGTTCCATTCGACTACACCCTGGCTGATGGCACTGAAGTCTCAGGTGCATGGGAGATGGAGGGG GACATGATGAAAGGCAAATTCACCAGGAAAGACAACAACAAGCTCCTGACCACTACTAGAACTCTGGTGGGCGGAGAGCT CGTGCAGAGCTACAACTATGAAGGGGTGGATGCCAAGAGGATTTTCAAGAAGCAGTAACCTGGATATTGAAGTTCAAGTT GTATTTTTATATAAATCACACAGCCCAGTGAACTGTATCTGAAGTTATTGTTGGCATTTAGCTTATTTCAGAAGAAGGAT CATAGTCCCCACGTTCATGAAACAACAGCTAGAAAAACACTGAAATGGTCACACAGCTCAGGTAAATTTGCATCCCTTTT CCTATGAAAGGAGAAAACAAACACTAAACAGCATCCTGGCAGTGGATGTCTATCTGTAGCAGTAGCTAAACATTCAAATG GGTTTAGTTTTATTTGTCTTGGAGGTGTTCAGGATAATCCGCACATCATAGTGTCAACAAATTTCACAGGACTCCTTCAT TGGTATGGTGTAGATTATCCACAGTAATGGGTGTTTTCAGGTAAAGCTCCACGGCAACATGAAATACATGAAATGCTATT GTGTGTGTTCAAACAGGGAAATAAATCAATAAATCAAATTGCACTAATGGTTCTCATGTATCAGATTGTCCTCTACAATA AAGCATGTTATGTCACTTGA
|
||||||||||