| Organism | |||
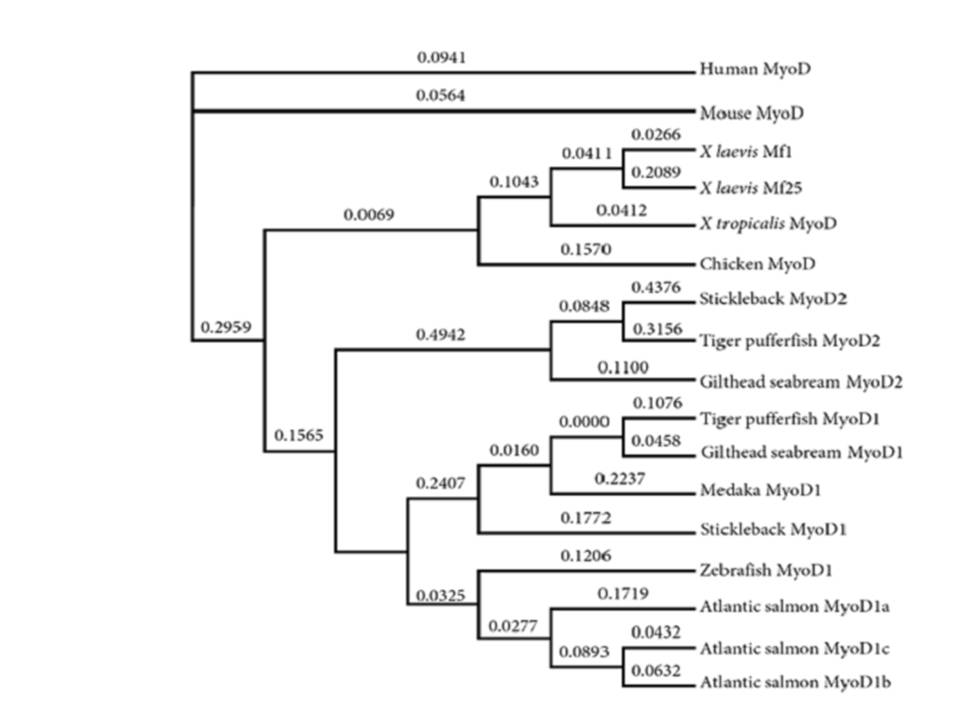
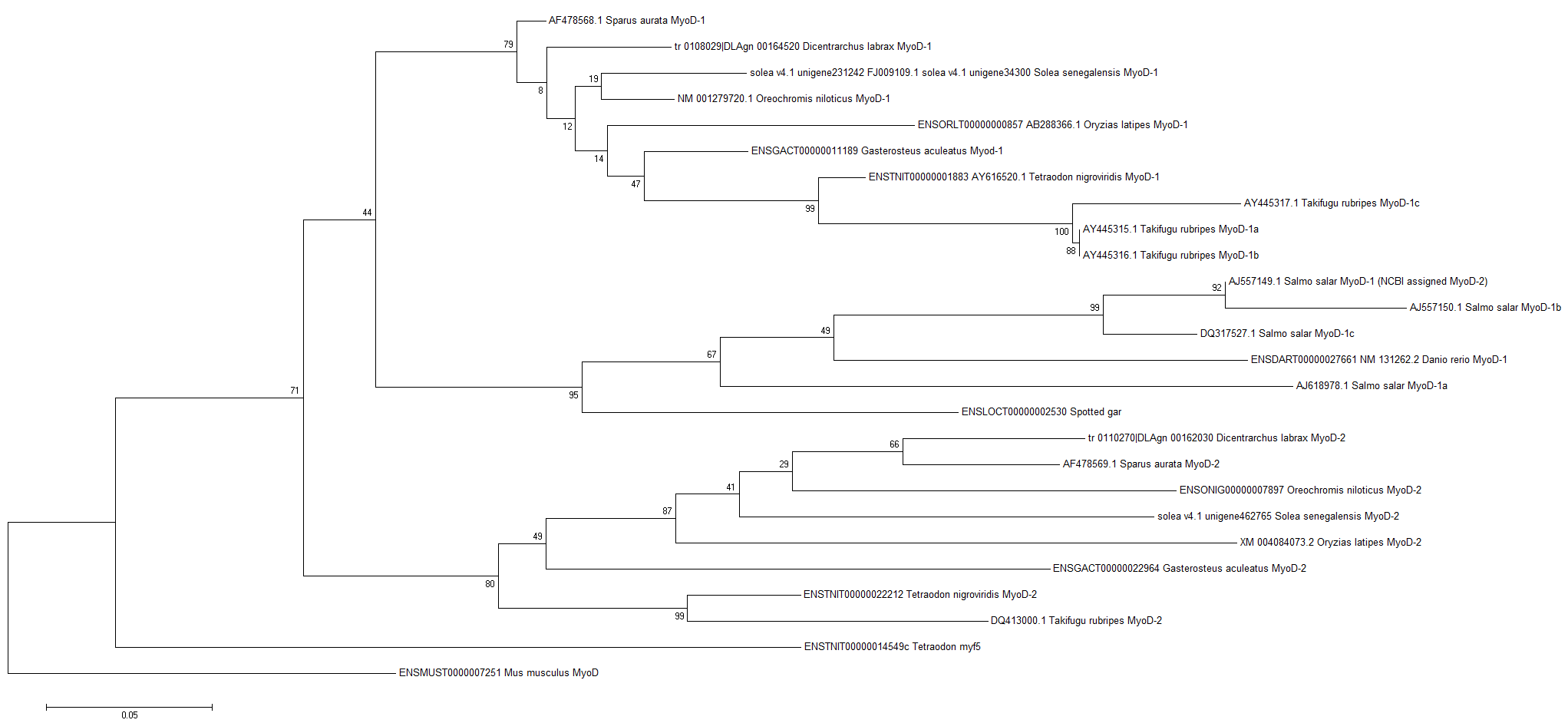
| Phylogeny | |||
| Genomic Map Locations | Dr_Chr.25 |
||
| Paralogs | |||
| Citations | |||
| Sequence | >NM_131262.2 Danio rerio Myogenic differentiation-1 CAAAGAAACCCTCCGAGGTCCTGCGAATTTTAGACTGTAGTTTTGAAACGTACGTTATTTCGGGATCTGAAGGACTTTGT CTTACTATTCCTAATTTTGGTTTTTATTTTTTTTACAAATTTACTACTTTGGAAACATTCAAGTACATTTCTTGCTTTTT AAACTTTAACACAAAAAAGATGGAGTTGTCGGATATCCCCTTCCCCATCCCATCAGCTGATGACTTCTACGACGACCCTT GCTTCAACACCAACGACATGCACTTCTTTGAAGACTTGGACCCCAGGCTTGTTCACGTGAGCCTGCTCAAACCCGACGAG CATCACCACATCGAGGACGAGCACGTGAGGGCGCCCAGTGGGCATCATCAGGCCGGCAGGTGCCTACTGTGGGCATGCAA AGCTTGCAAGAGAAAAACTACCAATGCTGACCGTCGCAAAGCCGCCACCATGAGGGAGAGGAGGCGACTGAGCAAGGTCA ACGACGCTTTCGAGACCCTCAAGAGATGCACGTCCACCAACCCGAACCAGAGGCTGCCCAAAGTGGAGATTCTGAGAAAC GCCATTAGTTATATCGAGTCTCTGCAGGCTCTTCTCAGAAGTCAAGAGGATAACTACTATCCCGTTCTGGAACATTACAG TGGAGACTCTGATGCTTCCAGTCCGAGATCCAACTGCTCTGATGGCATGATGGATTTTATGGGCCCAACGTGTCAGACGA GAAGACGGAACAGCTATGACAGCTCTTACTTCAATGACACACCAAATGCTGACGCACGGAATAATAAAAACTCAGTGGTG TCGAGTTTGGATTGTCTGTCCAGCATCGTGGAGCGAATTTCCACAGAGACTCCTGCATGTCCCGTGCTGTCAGTACCGGA GGGGCACGAAGAGAGCCCGTGTTCTCCGCATGAGGGATCTGTCCTGAGTGACACCGGAACCACCGCACCGTCCCCGACCA GCTGCCCTCAACAGCAGGCTCAGGAAACCATTTATCAAGTGCTTTAAAATTCTGCAACATTTCAAAACAAATTGAAAAAG ACAATCTGAATGAAGAAACTTTCAGCAGAAAAAGGCGAATCCGACCTTTAACGACAAAAGAAAGACTTTTGATCCACTGC TGGAAACTAGGAAAGAATGCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTC TTTCTTTCTTTCTTTCTTTCTTTCATAGACTAATTCTTTTTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTC TTTTTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTCTTTATGCGGTGGAAAA TGTAATTCCAATATGAGAAAAGACTCAGCAGATCAGCAGAAGTGAGTTTTTTAACTAAGCTTATTTATATTGTTATATCC ACGTGAAAATGTGACCATATTTTTTTCCCTTTGTGAATATATTTTCTGTCACCATCACCTTTTATTTTCCTAATTATTTA CGGAAAAAATCGTAATCCAGATACGAATAGTAGGACCATTTTTGTATATGTGTAAATAAGATCTGTTTTGTTAAAGCGAC GAAAACGAACAAACATATTATTGTATTTAATGATGCCTGTTTGAAACACTAGTTTGTTGTGTCTTGTGTTAAACTTTATA TTTATACTTCTTAAACGAGTGAATGACGGATAAATAAAAACAACTATTTATACTGGAAAAAAAAAAAAAAAAAAAAAAAA AA
|
||||||